In this case, Face2Gene helped Dr. Li target the analysis of a gene that otherwise would not have been analyzed, ultimately identifying the diagnostic variant.
WHO
Dr. Hong Li is a clinical and biochemical geneticist at Emory University specializing in the diagnosis and treatment of inherited metabolic disorders, lysosomal storage diseases, genetic disorders with dysmorphic features, and intellectual disability.
THE DILEMMA
For several years, the Emory Genetics team had been following a young female patient who remained undiagnosed. First seen by Genetics at age 5, the patient presented with global developmental delay, predominant speech delays, and microcephaly, but with otherwise normal growth. She also had early-onset epilepsy, starting at 1 year old, and was well-controlled on Keppra. Her MRI was essentially normal except for the left hippocampus with a globular configuration and abnormal folding pattern, but showed no abnormal signals or atrophy. Worried that there may be additional health issues on the way, the parents of this young girl were eager to get a diagnosis to prepare and provide the best care for their daughter. The patient’s neurologist ordered an NGS epilepsy gene panel, including analysis of over 100 genes before her consultation with Genetics. The panel returned several variants of uncertain significance and prompted various additional clinical work-ups in the pursuit of clinical correlation. However, none of these VUS results seemed to fit the patient’s clinical presentation.
When the patient returned to Genetics for her next visit at the age of 8, Dr. Li saw the patient and noticed some very unique facial features. She saw that the patient had hypotelorism and a slightly beaked nose. Although these features were not dramatic, they were uncommon and therefore caught Dr. Li’s attention.
THE SOLUTION
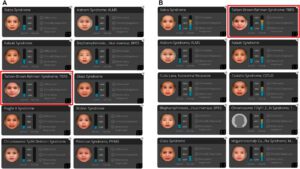
Having recently started using Face2Gene, Dr. Li decided to analyze this patient’s facial image. One of the top results returned was a syndrome associated with the DYRK1A gene, which was the second-ranked result and showed a moderate facial match. The fact that this syndrome was associated with her patient’s very unique features caught Dr. Li’s attention, and the Face2Gene heatmap helped to confirm these areas of strong facial similarity. Upon further review, Dr. Li was intrigued to see that the clinical features of this syndrome — including microcephaly, hypotelorism, pointed nose, and various neurological features — fit her patient’s clinical history very well.
She reached out to Dr. Lora Bean in the EGL Genetics lab, where the epilepsy panel had been completed, to see whether that panel had included an analysis of the DYRK1A gene. Though the gene had not been included in the panel, Dr. Bean was able to go back to the raw NGS data to look specifically at this gene.
THE RESULT
Sure enough, she found a suspicious variant. After analysis of parental samples, Dr. Bean confirmed that this was a de novo variant in the patient, which helped to classify this variant as “likely pathogenic” and causative for this patient’s clinical phenotypes.
The family of this young girl was relieved to have an answer, ending the diagnostic odyssey for their child. Finally, having a diagnosis allowed Dr. Li and the parents of this patient to set aside their worries about additional serious health issues.
Dr. Li continues to use Face2Gene to support her diagnostic evaluations, particularly when the patient has dysmorphic features. “Face2Gene is an integral part of my clinical evaluation,” said Dr. Li, “It’s a powerful tool that not only reaffirms my suspicion of a particular syndrome but has also pointed me towards syndromes I typically wouldn’t have considered, which have then resulted in the correct diagnosis.”
Dr. Li went on to add, “In this case, without the clues from Face2Gene, it was hard to consider any targeted testing due to the patient’s non-specific presentation. My next step would have been to order trio exome sequencing, but the integration of information from Face2Gene saved me this exome step.”