Personalizing Medicine with Artificial Intelligence and Facial Analysis
Presented by Omar Abdul-Rahman, MD, University of Nebraska Medical Center
Precision Medicine World Conference (PMWC)| September 24-25, 2018 | Duke University
This post is based on a presentation given at PMWC Duke. Watch the full presentation here.
At the Precision Medicine World Conference (PMWC) Duke meeting, Dr. Omar Abdul-Rahman, Friedland Professor and Director of Genetic Medicine at the University of Nebraska Medical Center, told the story of the emergence of phenotypic data as crucial in clinical evaluations.
According to Dr. Abdul-Rahman, there are, “three legs to a stool that we have to understand” when making a patient evaluation: genes, environment, and phenotype. He described a time about ten years ago when “there had been a lot of advancements made in the ability to get good genomic data.” However, he went on to say that those same advancements had unfortunately not been made in phenotyping.
Standardizing the Phenotype
The first step taken in tackling this standstill and beginning to capture robust phenotypic information was the development of the Elements of Morphology as a way to “standardize the nomenclature” of phenotypes. Together, a group of geneticists defined over 400 features of the face, hands, and feet to streamline they way clinicians referred to various morphologies.
“Once we standardized the phenotype, the next question became, ‘How do we capture it?’”
Capturing the Phenotype
While at a study site for the National Children’s Study, Dr. Abdul-Rahman and his team realized that they were successfully capturing genetic and environmental data, but not phenotypic. With the goal of capturing as many of the 400+ features as possible, they showed 15 photos (eight of the head/neck, four of the hands, and three of the feet) and three videos to a panel of geneticists for review.
This laborious study was presented as a poster at a conference where it was strategically placed next to a poster focused on computer-aided facial recognition. Combining Dr. Abdul-Rahman understanding of how to capture enough imaging to analyze a phenotype with his colleagues’ computer-aided automation of the process, a collaborative study between the brains behind the neighboring posters was born.
Analyzing the Phenotype
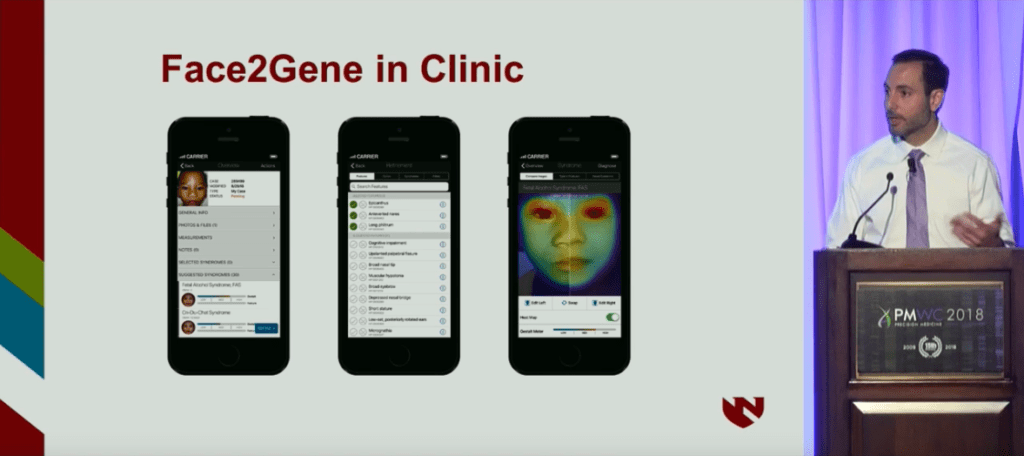
The automating software introduced, Face2Gene is a suit of phenotyping applications developed by FDNA that operates on AI and deep learning technologies. Using facial analysis, the system has evaluated over 150,000 patients, and cross-comparing their phenotypes to a growing database of over 10,000 genetic diseases helps to enable more rapid and accurate diagnoses.
Studying FASD with Face2Gene
After learning about Face2Gene, Dr. Rahman chose to apply the technology to a particular teratogen of interest, alcohol, and related Fetal Alcohol Spectrum Disorders (FASD). FASD can be broken up into four diagnostic categories, the first three of which are straightforward:
- FAS – requires all of the typically-present features
- Partial FAS – requires some, but not all of the typically-present features
- Alcohol-related birth defects (ARBD) – includes a series of anomalies more common to present among alcohol consumers
- Alcohol-related neurodevelopmental disorder (ARND)
The final FASD category, Alcohol-related neurodevelopmental disorder (ARND), is more difficult to diagnose because there are no outward features. When diagnosing, there are two requirements: documented prenatal alcohol consumption and neurobehavior impairment, which cannot be diagnosed under the age of three.
Because ARND is both the most difficult FASD to diagnose and also the most prevalent FASD type in the United States, Dr. Abdul-Rahman decided to put Face2Gene to the test to see if the technology could detect the difference between the four diagnostic categories. He ran images of over 130 subjects with FAS, Partial FAS, ARBD, or ARND, as well as controls through Face2Gene in a cross-validation test. The images were split 50/50 into training and test sets and were run through ten rounds. The results showed that for any FASD vs. control, the manual and computer-aided score both performed well and relatively equally. The same was true of looking at the individual syndrome categories, FAS, Partial FAS, and ARBD; however, when looking at ARND vs. control, there was a better AUC for computer-aided vs. manual.
You can find the study here.
Lessons Learned
Although there appeared to be no physical characteristics in the clinical criteria for ARND, Face2Gene was able to pick up on subtle facial cues, showing that, “there must be something that is broader, but is really subclinical from a provider standpoint.” Dr. Abdul-Rahman concluded with the following lessons learned from the study and his experience using Face2Gene:
- Facial analysis software like FDNA’s Face2Gene can be used to delineate phenotypes at a subclinical level
- Such software may allow for the identification of external physical biomarkers for conditions such as FASD
- Such software can potentially be used by primary care providers as a screen for further investigations (e.g. genetics referral)
Check out other presentations and similar research on the benefits of AI and facial analysis in personalized medicine here.